3.3. Perl modules#
For installing Perl modules, you can configure
the local-lib module to create and use a local
directory. The default location is $HOME/perl5.
If your software of interest is available on CPAN, you can install it easily using the cpanminus script.
Note
Your software of interest, might be available using The quick and easy method.
Search for package names containing perl.
3.3.1. Requirements#
The build-essential and cpanminus packages installed#
If you do not have them installed, follow The quick and easy method, and search and install the following packages:
build-essential— installs compilers and build tools like
make
cpanminus— script to obtain, build and install modules from CPAN along with their dependencies
Attention
You should not add sudo in the commands below.
This method only installs files in your home directory and so does not require administrator privileges.
3.3.2. Configuring local-lib module#
To configure local-lib:
Open the $HOME/.bashrc file in a text editor.
Add the following text at the end of the file:
# Configuration for Perl local-lib module
eval "$(perl -Mlocal::lib)"
Save the file.
Here is a sample .bashrc, for reference
(Fig. 57).
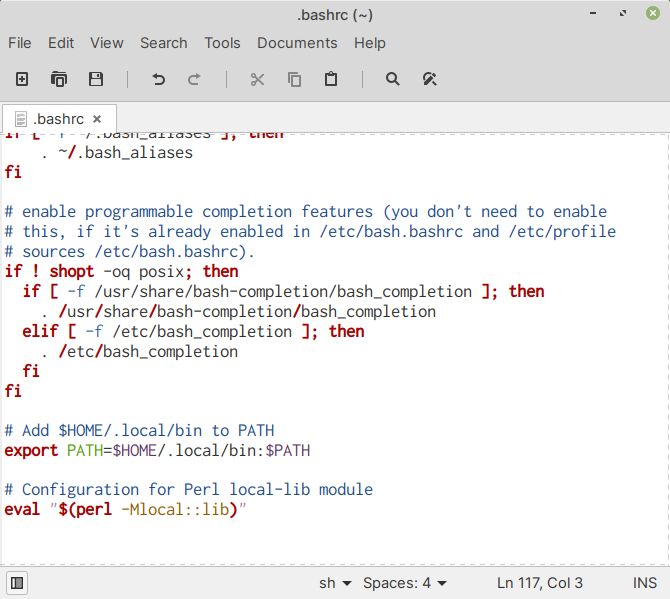
Fig. 57 local-lib configuration in .bashrc#
Open a new terminal window.
You will notice the following message displayed (Fig. 58):
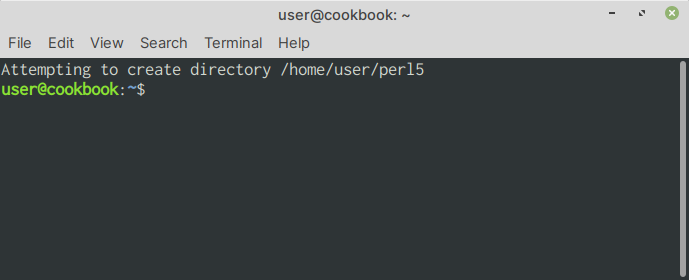
Fig. 58 Attempting to create directory /home/user/perl5#
Configuration is now complete.
You can now start installing modules from CPAN and other sources.
3.3.3. Searching for a module on CPAN#
Open MetaCPAN website (https://metacpan.org) in a web browser.
In the Search field, enter the name of the module you would like to install and click on the Search the CPAN button.
I will use David H. Ardell’s FAST package as an example. (Fig. 59).

Fig. 59 Searching for a module on MetaCPAN#
This should display modules matching the search term —
FAST (Fig. 60).
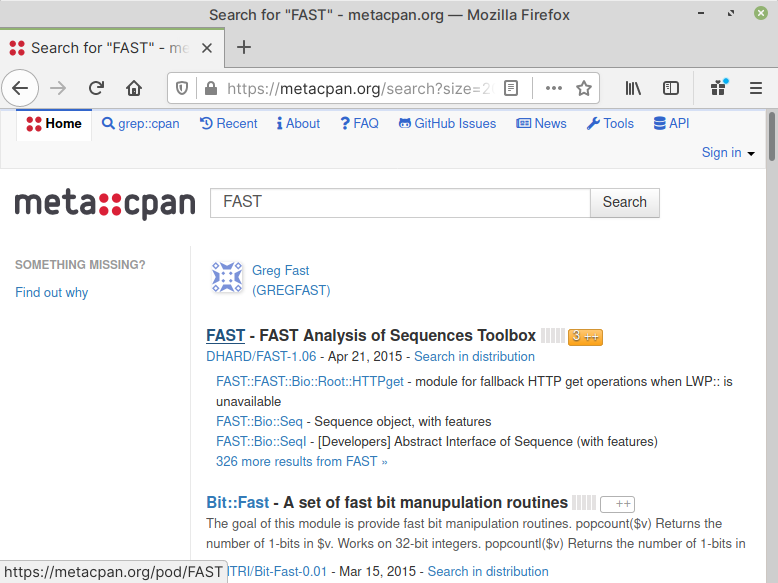
Fig. 60 Search results for modules#
If you click on the appropriate module name, you will be taken to a description page, where you can learn more about the module (Fig. 61).
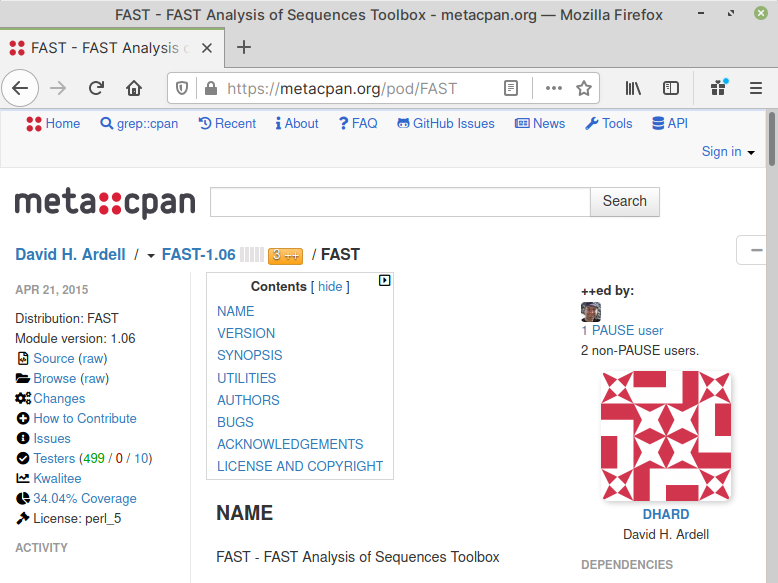
Fig. 61 Module description page#
On this page, you can also verify the module name to use in the next step.
To do that, click on Install Instructions
under TOOLS menu in the sidebar. This will
open a pop-up window with information on the module
name you need to use with the cpanm command.
In this case, the module name to use, is simply
called FAST (Fig. 62).
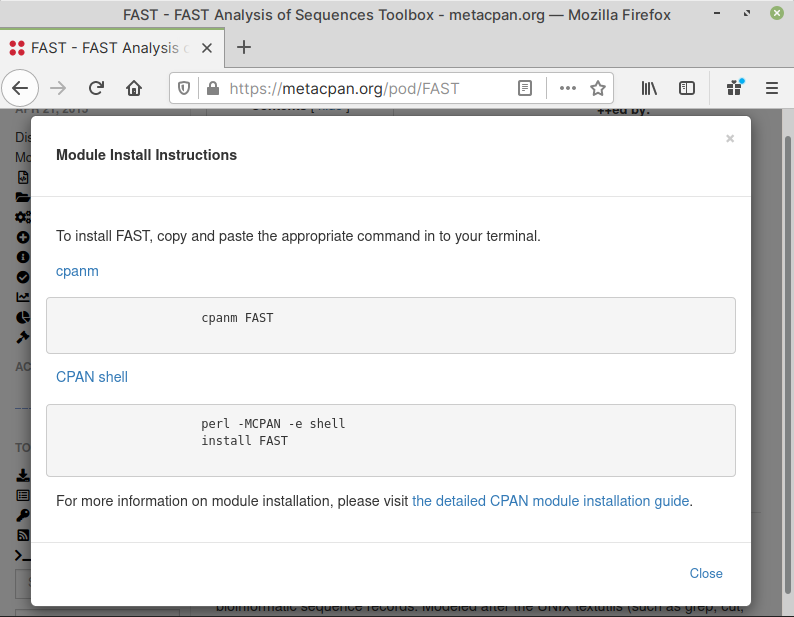
Fig. 62 Module install instructions for FAST#
You can now proceed towards installing this module.
3.3.4. Installing a module#
Open a terminal window.
Install the module with the cpanm command:
cpanm FAST
This will install the module and its dependencies, if there are any.
Output:
--> Working on FAST
Fetching http://www.cpan.org/authors/id/D/DH/DHARD/FAST-1.06.tar.gz ... OK
Configuring FAST-1.06 ... OK
==> Found dependencies: Bit::Vector, Sort::MergeSort, Sort::Key
--> Working on Bit::Vector
Fetching http://www.cpan.org/authors/id/S/ST/STBEY/Bit-Vector-7.4.tar.gz ... OK
Configuring Bit-Vector-7.4 ... OK
...
Building and testing FAST-1.06 ... OK
Successfully installed FAST-1.06
6 distributions installed
3.3.5. Using installed modules#
Using included commands#
If the module includes commands, those will be installed
in $HOME/perl5/bin.
This directory will be included in $PATH, when you
configured local-lib.
Hence, you can start using the commands included in a
module immediately after installation.
For example, here is the help page of the fasuniq
command included with the FAST package installed in the
previous step:
fasuniq -h
Usage:
fasuniq [options] [MULTIFASTA-FILE]
[MULTIFASTA-DATA-ON-STDIN] | fasuniq [options]
...
Using modules in your scripts#
Modules will be installed in:
$HOME/perl5/lib/perl5/5.30.0.
Note
This is the module path for Perl 5.30.0.
It you have a different version of Perl installed on your system, it will change.
As local-lib makes these modules discoverable automatically, you can start using them in your scripts immediately.
Here is an example using the Bio::Phylo package.
After installing the module with cpanm Bio::Phylo,
create a file using a text editor and copy the following
content:
use Bio::Phylo;
# print version
print Bio::Phylo->VERSION;
# print citation
print Bio::Phylo->CITATION;
Save the file as biophylo_test.pl and then run it:
perl biophylo_test.pl
Output:
v2.0.1Rutger A Vos, Jason Caravas, Klaas Hartmann, Mark A Jensen and Chase Miller, 2011.
Bio::Phylo - phyloinformatic analysis using Perl. BMC Bioinformatics 12:63.
doi:10.1186/1471-2105-12-63
Comments